Abstract
Proteasome inhibitors were first introduced to the clinic almost 20 years ago and have since become standard of care in multiple myeloma treatment, a cancer of terminally differentiated plasma cells. The proteasome degrades most cellular proteins and identifies targets following ubiquitination by an elaborate enzymatic cascade. Blocking the turnover of proteins with proteasome inhibitors affects many pathways, including signaling, metabolism, and stress responses. Transcriptional and epigenetic regulators are short-lived proteins, and proteasome inhibition is expected to alter gene activity dramatically. However, some of the least understood aspects of proteasome inhibitors involve their effects on epigenetics and transcription. One reason for this knowledge gap is the technical challenge of distinguishing the direct from indirect effects of proteasome inhibition on transcription.
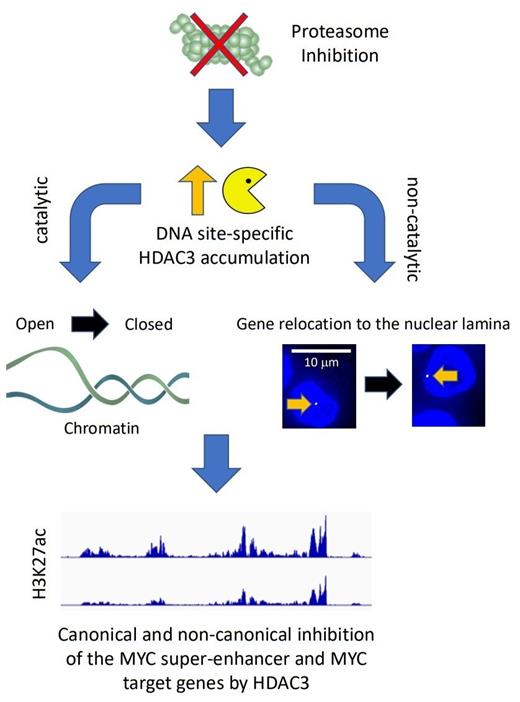
To overcome this limitation, we developed an approach to map the nuclear location of protein turnover. We detected MYC target genes as a prevalent site of proteasomal protein degradation in multiple myeloma cells. Multiple myeloma is addicted to the proto-oncogene MYC, and several new approaches are being tested to silence MYC in this cancer. MYC is a short-lived protein, and proteasome inhibitors should stabilize MYC. However, with increased MYC levels, it is unclear why proteasome inhibition would be clinically beneficial. Instead, we found that proteasome inhibition reduces the levels of MYC and the activity of its target genes. The surprising reduction of MYC by proteasome inhibitors makes sense given their clinical effect. To identify the molecular mechanism by which proteasome inhibitors suppress MYC, we performed an integrative genomic analysis on the effects of these drugs and found that MYC is silenced at the transcriptional level by epigenetic suppression of its super-enhancer.
We found that acetylated H3K27, a histone modification that increases the accessibility of chromatin and facilitates transcription, is rapidly lost upon proteasome inhibition. We hypothesized that this is caused by the stabilization of a histone deacetylase (HDAC). Based on data from the APEX study, we discovered that HDAC3 antagonizes the activity of MYC, and cancers with high HDAC3 expression correlate with better outcomes. Indeed, we found that proteasome inhibition locally increases HDAC3 levels at target promoters and the MYC super-enhancer and that genetic depletion of HDAC3 reduces the epigenetic effects of proteasome inhibition. In addition to its epigenetic role, HDAC3 has recently been shown to repress genes by disrupting their location. In agreement with these findings, we discovered that target genes relocate into heterochromatin-rich lamina-associated areas of the nucleus upon proteasome inhibition. These results suggest that HDAC3 might also act as a suppressor of MYC in a manner that does not require catalytic deacetylase activity. Such a repressive function would, therefore, not be targeted by HDAC inhibitors.
In summary, our study supports a new role of proteasome inhibitors as antagonists of the proto-oncogene MYC. The drugs accomplish this effect by stabilizing HDAC3 at the MYC super-enhancer and MYC target genes. As a consequence, elevated HDAC3 represses chromatin epigenetically and possibly alters the nuclear architecture by relocating chromosomes. These findings are surprising as they point towards a novel mechanism to limit MYC, which is dysregulated in 70% of cancers. Our results may also explain why some highly MYC-addicted cancers have a particular sensitivity towards proteasome inhibitors.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal